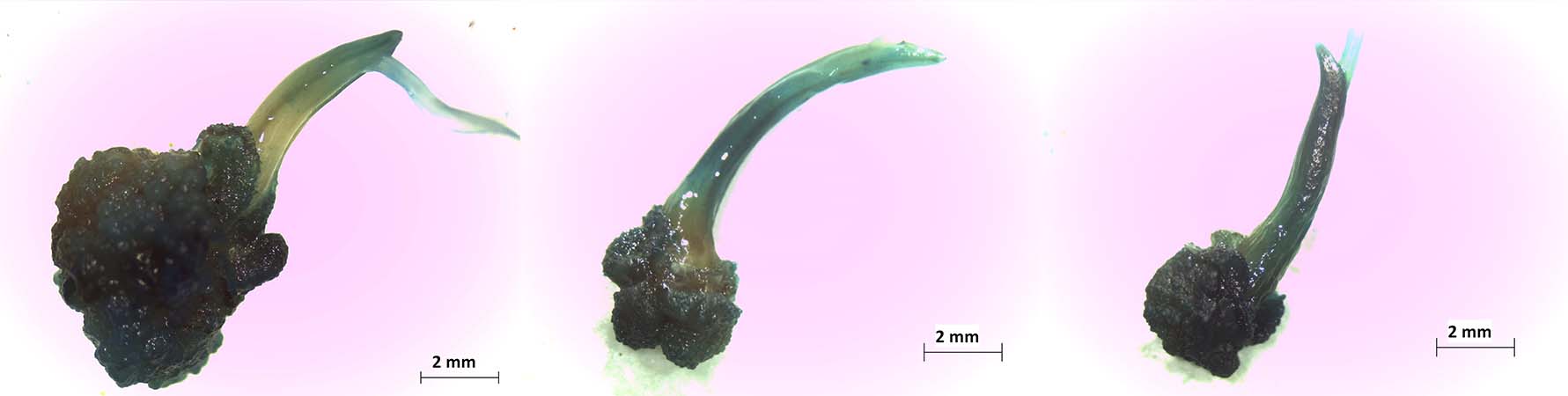
Key aspects for the genetic transformation of rice (Oryza sativa L.) subspecies indica by Agrobacterium tumefaciens
DOI:
https://doi.org/10.15517/am.v32i3.44978Keywords:
β-glucuronidase, gus, genetic modification, embryogenic callusAbstract
Introduction. Agrobacterium-mediated transformation of rice (Oryza sativa L. ssp indica) represents an opportunity for scientific research and genetic improvement. Optimization of the protocol is necessary to obtain the highest transformation efficiency. Objective. To evaluate different factors that affect the genetic transformation in embryogenic rice callus of subspecies indica through Agrobacterium tumefaciens. Materials and methods. This study was performed in San José, Costa Rica between 2012 and 2014. The following were evaluated in six treatments: the effect of callus age, acetosyringone concentration, lighting condition, the presence or absence of radicle, and Agrobacterium tumefaciens strain on the genetic transformation of embryogenic calli of rice variety CR5272 with gus reporter gene. Agrobacterium strains LBA4404 with pCAMBIA1305.2 plasmid, and strains ATHV, GV3101, and LBA4404 with the pCAMBIA 1303 plasmid were compared; by histochemical tests for the detection of transient expression of the β-glucuronidase reporter gene. Results. The evaluation of the six treatments with strain LBA4404::pCAMBIA 1305.2 resulted in transient expression of 1.33-7.00 % of the GusPlus gene for the CR5272 variety, and 8.00 % for the control with the Nipponbare (ssp. japonica) variety. The strains with the pCAMBIA1303 plasmid showed a transient expression of the gusA gene between 100-65 % with an average area of 14.23 mm2 (ATHV), 8.81 mm2 (GV3101), and 8.83 mm2 (LBA4404) with no significant differences between them; however, there were differences when compared with strain LBA4404::pCAMBIA1305.2 (85 %, 4.39 mm2). Conclusions. The use of the conditions: six-day callus, acetosyringone concentration of 76 µM, light before and after cocultivation, presence of radicle and the ATHV::pCAMBIA 1303 , improved the transformation efficiency with Agrobacterium tumefaciens in the rice variety CR5272.
Downloads
References
Abdollahi, M. R., Memari, H. R., & van Wijnen, A. J. (2011). Factor affecting the endogenous β-glucuronidase activity in rapeseed haploid cells: how to avoid interference with the Gus transgene in transformation studies. Gene, 487, 96–102. https://doi.org/10.1016/j.gene.2011.07.007
Alok, A., Sharma, S., Kumar, J., Verma, S., & Sood, H. (2017). Engineering in plant genome using Agrobacterium: progress and future. In V. C. Kalia, & A. K. Saini (Eds.), Metabolic engineering for bioactive compounds: Strategies and processes (pp. 91–111). Springer. https://doi.org/10.1007/978-981-10-5511-9_5
Bai, C., Rivera, S., Medina, V., Alves, R., Vilaprinyo, E., Sorribas, A., Canela, R., Capell, T., Sandmann, G., Christou, P., & Zhu, C. H. (2014). An in vitro system for the rapid functional characterization of genes involved in carotenoid biosynthesis and accumulation. The Plant Journal, 77, 464–475. https://doi.org/10.1111/tpj.12384
Bonilla, M., Muñoz, J., & Sánchez, F. (2008). Expresión transitoria del gen gus en caña de azúcar usando Agrobacterium tumefaciens. Acta Agronómica, 57(3), 161–166.
Camacho, J., & Navarro, J. (2020). Selección de líneas promisorias de arroz a partir de generaciones avanzadas. Alcances Tecnológicos, 13(1), 40–49. https://doi.org/10.35486/at.v13i1.169
Cervera, M. (2005). Histoquemical and fluorometric assays for uidA (GUS) gene detection. In L. Peña (Ed.), Methods in molecular biology, transgenic plants: Methods and protocols (pp. 203–211). Humana Press Inc. https://doi.org/10.1385/1-59259-827-7:203
Cheng, X., Sardana, R., Kaplan, H., & Altosaar, I. (1998). Agrobacterium-transformed rice plants expressing synthetic cryIA (b) and cry IA (c) genes are highly toxic to striped stem borer and yellow stem borer. Applied Biological Sciences, 95, 2767–2772. https://doi.org/10.1073/pnas.95.6.2767
Corporación Arrocera Nacional. (2019). Informe estadístico periodo 2018-2019. https://www.conarroz.com/userfile/file/INFORME_ANUAL_ESTADISTICO_PERIODO_2018_2019.pdf
Dai, S., Zhenng, P., Marmey, P., Zhang, S., Tian, W., Chen, S., Beachy, R. N., & Fauquet, C. (2001). Comparative analysis of transgenic rice plants obtained by Agrobacterium-mediated transformation and particle bombardment. Molecular Breeding, 7, 25–33. https://doi.org/10.1023/A:1009687511633
Datta, K., & Datta, S. K. (2006). Indica rice (Oryza sativa, BR29 and IR64). In K. Wang (Ed.), Agrobacterium Protocols. Methods in Molecular Biology (pp. 201–212). Humana Press Inc. https://doi.org/10.1385/1597451304
Deeba, F., Hyder, M. Z., Shah, S. H., & Naqvi, S. M. (2014). Multiplex PCR assay for identification of commonly used disarmed Agrobacterium tumefaciens strains. SpringerPlus, 3, Article 358. https://doi.org/10.1186/2193-1801-3-358
Food and Agriculture Organization. (2004). Rice and human nutrition. Food and Agriculture Organization.
Fukagawa, N. K, & Ziska, L. H. (2019). Rice: Importance for global nutrition. Journal of Nutritional Science and Vitaminology, 65, S2–S3. https://doi.org/10.3177/jnsv.65.S2
García-Arias, C. (2011). Transformación del genoma de arroz (Oryza sativa L.) con el gen Vip3A de Bacillus thuringiensis para conferir tolerancia a Spodoptera frugiperda (J.E SMITH) [Tesis de Maestría, no publicada]. Universidad de Costa Rica.
He, X., Batheja, M., & Fuqua, C. (2005). Promoter-probe cassettes with the gusA (β-glucoronidase) reporter gene and several different antibiotic resistance markers. Journal of Microbiological Methods, 60, 281–283. https://doi.org/10.1016/j.mimet.2004.10.005
Hiei, Y., & Komari, T. (2006). Improved protocols for transformation of indica rice mediated by Agrobacterium tumefaciens. Plant Cell, Tissue and Organ Culture, 85, 271–283. https://doi.org/10.1007/s11240-005-9069-8
Hiei, Y., Ohta, S., Komari, T., & Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant Journal, 6(2), 271–282. https://doi.org/10.1046/j.1365-313x.1994.6020271.x
Hoque, M. E., Mansfield, J. W., & Bennett, M. H. (2005). Agrobacterium-mediated transformation of indica rice genotypes: an assessment of factors affecting the transformation efficiency. Plant Cell, Tissue and Organ Culture, 82, 45–55. https://doi.org/10.1007/s11240-004-6154-3
Jefferson, R. A., Burgess, S.M., & Hirsh, D. (1986). β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proceedings of the National Academy of Sciences, 83(22), 8447–8451. https://doi.org/10.1073/pnas.83.22.8447
Kosugi, S., Ohashi, Y., Nakajima, K., & Arai, Y. (1990). An improved assay for β-glucuronidase in transformed cells: Methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Science, 70, 133–140. https://doi.org/10.1016/0168-9452(90)90042-M
Kumar, K. K., Maruthasalam, S., Loganathan, M., Sudhakar, D., & Balasubramanian, P. (2005). An improved Agrobacterium mediated transformation protocol for recalcitrant elite indica rice cultivars. Plant Molecular Biology Reporter, 23, 67–73. https://doi.org/10.1007/BF02772648
Kuta, D., & Tripathi, L. (2005). Agrobacterium- induced hypersensitive necrotic reaction in plant cells: a resistance response against Agrobacterium-mediated DNA transfer. African Journal of Biotechnology, 4, 752-757.
León, J., & Arroyo, N. (2011). Producción, tecnología y comercialización del arroz en Costa Rica 1950-2005. Universidad de Costa Rica.
Lin, Y., & Zhang, Q. (2005). Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Reports, 23, 540–547. https://doi.org/10.1007/s00299-004-0843-6
Murashige, T., & Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum, 15, 473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Nishimura, A., Aichi, I., & Matsuoka, M. (2006). A protocol for Agrobacterium-mediated transformation in rice. Nature protocols, 1(6), 2796-2802. https://doi.org/10.1038/nprot.2006.469
Opabode, J. T. (2006). Agrobacterium-mediated transformation of plants: emerging factors that influence efficiency. Biotechnology and Molecular Biology Review, 1(1), 12–20.
Patel, M., Dewey, R. E., & Qu, R. (2013). Enhancing Agrobacterium tumefaciens-mediated transformation efficiency of perennial ryegrass and rice using heat and high maltose treatments during bacterial infection. Plant Cell, Tissue and Organ Culture, 114, 19–29. https://doi.org/10.1007/s11240-013-0301-7
R Core Team. (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org.
Ramesh, S., Nagadhara, D., Reddy, V. D., & Rao, K. V. (2004). Production of transgenic indica rice resistant to yellow stem borer and sap-sucking insects, using super-binary vectors of Agrobacterium tumefaciens. Plant Science, 166, 1077–1085. https://doi.org/10.1016/j.plantsci.2003.12.028
Ramesh, M., & Gupta, A. (2005). Transient expression of beta-glucuronidase gene in indica and japonica rice (Oryza sativa L.) callus cultures after diferent stages of co-bombardment. African Journal of Biotechnology, 4(7), 596–600. https://doi.org/10.5897/AJB2005.000-3108
Reddy, V. S. (2008). Meetings and courses: Theoretical and practical course “Transgene expression in plants”. International Centre for Genetic Engineering and Biotechnology.
Saharan, V., Yadav, R. C., Yadav, N. R., & Ram, K. (2004). Studies on improved Agrobacterium-mediated transformation in two indica rice (Oryza sativa L.). African Journal of Biotechnology, 3(11), 572–575. https://doi.org/10.4314/ajb.v3i11.15020
Saikat, P., & Aryadeep, R. (2019). Comparative analyses of regeneration potentiality of eight indigenous aromatic indica rice (Oryza sativa L.) varieties. International Journal of Scientific Research in Biological Sciences, 6(1), 55–64. https://doi.org/10.26438/ijsrbs/v6i1.5564
Sawant, G. B., Sawardekar, S. V., Bhave, S. G., & Kshirsagar, J. K. (2018). Effect of acetosyringone and age of callus on Agrobacterium - mediated transformation of rice (Oryza sativa L.) calli. International Journal of Chemical Studies, 6(3), 82–88.
Schneider, C. A., Rasband, W. S., & Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9(7), 671–675. https://doi.org/10.1038/nmeth.2089
Shri, M., Rai, A., Verma, P. K., Misra, P., Dubey, S., Kumar, S., Verma, S., Gautam, N., Tripathi, R., Trivedi, P., & Chakrabarty, D. (2013). An improved Agrobacterium-mediated transformation of recalcitrant indica rice (Oryza sativa L.) cultivars. Protoplasma, 250, 631–636. https://doi.org/10.1007/s00709-012-0439-x
Sood, P., Bhattacharya, A., & Sood, A. (2011). Problems and possibilities of monocot transformation. Biología plantarum, 55(1), 1–15. https://doi.org/10.1007/s10535-011-0001-2
Sudan, C., Prakash, S., Bhomkar, P., Jain, S., & Bhalla-Sarin, N. (2006). Ubiquitous presence of β-glucuronidase (gus) in plants and its regulation in some model plants. Planta, 224(4), 853–864. https://doi.org/10.1007/s00425-006-0276-2
Sundararajan, S., Sivaraman, B., Rajendran, V., & Ramalingam, S. (2017). Tissue culture and Agrobacterium- mediated genetic transformation studies in four commercially important indica rice cultivars. Journal of Crop Science and Biotechnology, 20(3), 175–183. https://doi.org/10.1007/s12892-017-0045-0
Tan, L. W., Rahman, Z. A., Goh, H. H., Hwang, D. J., Ismail, I., & Zainal, Z. (2017). Production of transgenic rice (indica cv. MR219) overexpressing ABP57 gene through Agrobacterium-mediated transformation. Sains Malaysiana, 46(5), 703–711. https://doi.org/10.17576/jsm-2017-4605-04
Tie, W., Zhou, F., Wang, L., Xie, W., Chen, H., Li, X., & Lin, Y. (2012). Reasons for lower transformation efficiency in indica rice using Agrobacterium tumefaciens-mediated transformation: lessons from transformation assays and genome-wide expression profiling. Plant Molecular Biology, 78, 1–18. https://doi.org/10.1007/s11103-011-9842-5
Toki, S. (1997). Rapid and efficient Agrobacterium mediated transformation in rice. Plant Molecular Biology Reporter, 15(1), 16–21. https://doi.org/10.1007/BF02772109
Toriyama, K., & Hinata, K. (1985). Cell suspension and protoplast culture in rice. Plant Science, 41, 179–183. https://doi.org/10.1016/0168-9452(85)90086-X
Tripathi, R. M., Bisht, H. S., & Singh, R. P. (2010). Effect of acetosiringone and callus age on transformation for scutellum-derived callus of rice. International Journal of Pharma and Bio Sciences, 1(4), 163–171.
Tzfira, T., Jensen, C.S, Wang, W., Zuker, A., Vinocur, B., Altman, A., & Vainstein, A. (1997). Transgenic Populus tremula: a step-by-step protocol for its Agrobacterium- mediated transformation. Plant Molecular Biology Reporter, 15(3), 219–235.
United Nations. (2019). (2020, January 7). World population prospects. https://population.un.org/wpp/
Valdés, V., Aguilar, J., & Sanabria, A. (1992). Tecnología de producción para el cultivo de arroz en riego: Mejores alternativas en el distrito de riego Arenal (Cuaderno informativo para productores No. 2). MAG-SENARA-IICA.
Vega, R., Vásquez, N., Espinoza, A. M., Gatica, A., & Valdez-Melara, M. (2009). Histology of somatic embryogenesis in rice (Oryza sativa cv. 5272). Revista Biología Tropical, 57(1), 141–150.
Yookongkaew, N., Srivatanakul, M., & Narangajavana, J. (2007). Development of genotype-independent regeneration system for transformation of rice (Oryza sativa ssp. indica). Journal of Plant Research, 120(2), 237–245.
Downloads
Additional Files
Published
How to Cite
Issue
Section
License
1. Proposed policy for open access journals
Authors who publish in this journal accept the following conditions:
a. Authors retain the copyright and assign to the journal the right to the first publication, with the work registered under the attribution, non-commercial and no-derivative license from Creative Commons, which allows third parties to use what has been published as long as they mention the authorship of the work and upon first publication in this journal, the work may not be used for commercial purposes and the publications may not be used to remix, transform or create another work.
b. Authors may enter into additional independent contractual arrangements for the non-exclusive distribution of the version of the article published in this journal (e.g., including it in an institutional repository or publishing it in a book) provided that they clearly indicate that the work was first published in this journal.
c. Authors are permitted and encouraged to publish their work on the Internet (e.g. on institutional or personal pages) before and during the review and publication process, as it may lead to productive exchanges and faster and wider dissemination of published work (see The Effect of Open Access).